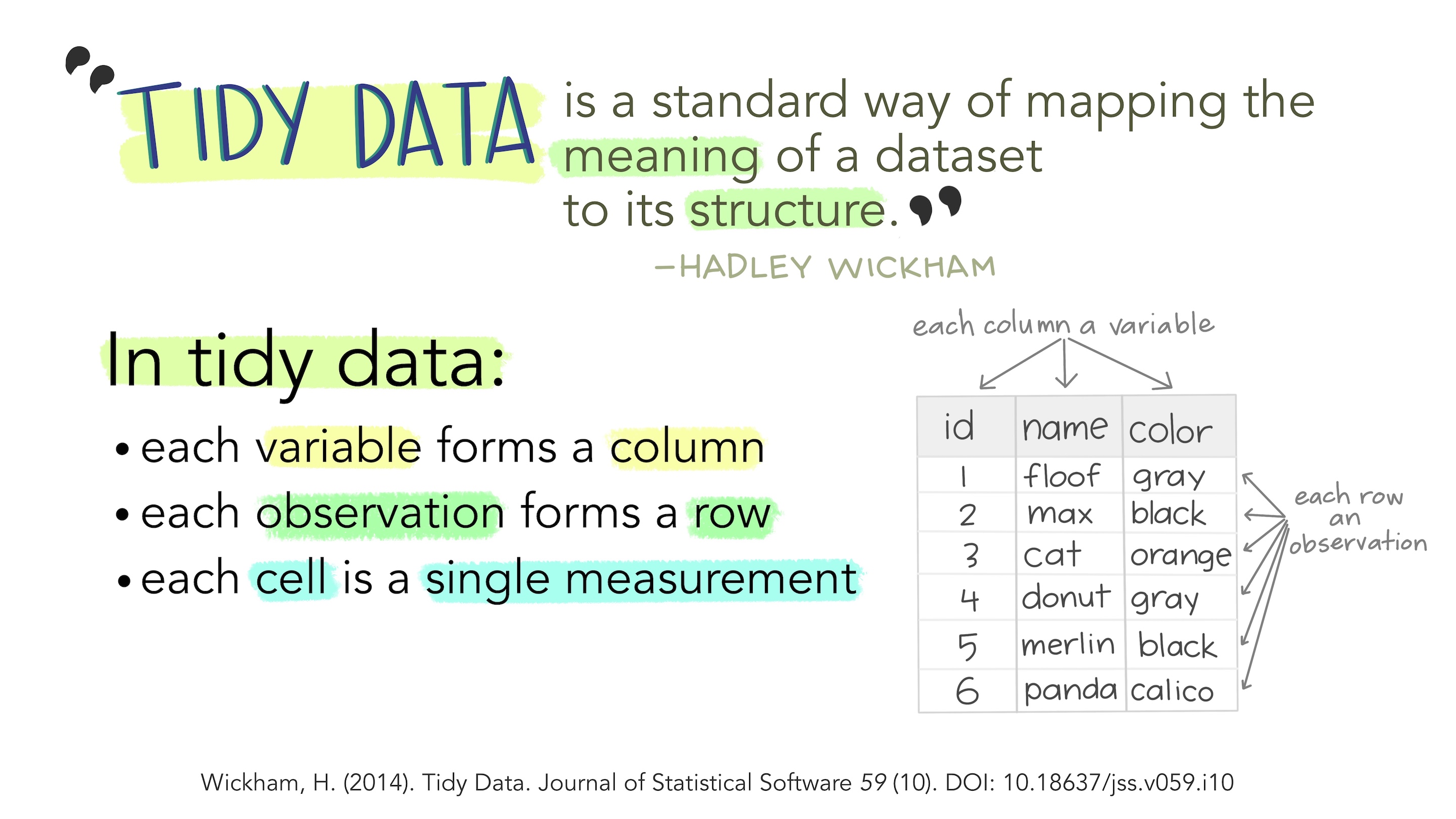
Rows: 344
Columns: 17
$ studyName <chr> "PAL0708", "PAL0708", "PAL0708", "PAL0708", "PAL…
$ `Sample Number` <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 1…
$ Species <chr> "Adelie Penguin (Pygoscelis adeliae)", "Adelie P…
$ Region <chr> "Anvers", "Anvers", "Anvers", "Anvers", "Anvers"…
$ Island <chr> "Torgersen", "Torgersen", "Torgersen", "Torgerse…
$ Stage <chr> "Adult, 1 Egg Stage", "Adult, 1 Egg Stage", "Adu…
$ `Individual ID` <chr> "N1A1", "N1A2", "N2A1", "N2A2", "N3A1", "N3A2", …
$ `Clutch Completion` <chr> "Yes", "Yes", "Yes", "Yes", "Yes", "Yes", "No", …
$ `Date Egg` <date> 2007-11-11, 2007-11-11, 2007-11-16, 2007-11-16,…
$ `Culmen Length (mm)` <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34…
$ `Culmen Depth (mm)` <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18…
$ `Flipper Length (mm)` <dbl> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190,…
$ `Body Mass (g)` <dbl> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 34…
$ Sex <chr> "MALE", "FEMALE", "FEMALE", NA, "FEMALE", "MALE"…
$ `Delta 15 N (o/oo)` <dbl> NA, 8.94956, 8.36821, NA, 8.76651, 8.66496, 9.18…
$ `Delta 13 C (o/oo)` <dbl> NA, -24.69454, -25.33302, NA, -25.32426, -25.298…
$ Comments <chr> "Not enough blood for isotopes.", NA, NA, "Adult…